The time-course and RNA interference of TNF-α, IL-6, and IL-1β expression on neuropathic pain induced by L5 spinal nerve transection in rats
Article information
Abstract
Background
The objective of this study was to investigate the time-course of the expression of TNF-α, IL-6, and IL-1β after L5 spinal nerve transection (SNT), and to determine the effect of small interfering RNA (siRNA) targeting these cytokines on neuropathic pain.
Methods
Rats received control siRNA (CON group, n = 80) or a cocktail of siRNAs targeting these cytokines (COCK group, n = 70). The siRNAs were given via intrathecal catheter 1 d prior to SNT, on the operation day, and 1, 2 and 3 d postoperatively. Behavioral tests and levels of the cytokine mRNAs and proteins as well as glial cell activity were following the L5 SNT.
Results
In the CON group, TNF-α and IL-1β mRNA levels increased immediately after SNT and remained high for 6 d, while IL-6 transcripts only began to increase after 12 h. TNF-α and IL-1β mRNA levels in the COCK group were lower than in the CON group at all time points (P < 0.05). In the behavioral tests, allodynia and hyperalgesia were significantly lower in the COCK group from 2 d after SNT (P < 0.05).
Conclusions
The time courses of TNF-α, IL-6 and IL-1β mRNA expression after L5 SNT differ. RNA interference may be a method of reducing the development of mechanical allodynia and hyperalgesia in response to nerve injury.
Introduction
Neuropathic pain is a pathological pain that persists despite resolution of the original damage, and is characterized by augmented responses to noxious stimuli (hyperalgesia), pain elicited by stimuli which do not normally provoke pain (allodynia) and spontaneous pain [1]. In addition to neurons, non-neuronal cells, such as glia, play a role in the development of neuropathic pain. Cytokines secreted by glial cells of the central nervous system, such as TNF-α, IL-6 and IL-1β, are involved in the development of neuropathic pain [2]. In animal studies, endoneural injection of TNF-α produced allodynia and hyperalgesia, while blocking TNF-α receptors reduced behavior related to neuropathic pain [2]. IL-6-mediated hyperalgesia/allodynia has also been reported in a rat mononeuropathy model [3], while IL-1β is a crucial cytokine secreted in response to nerve injury, and intrathecal administration of IL-1β induced hyperalgesia in rats [4]. Although it has been shown that these cytokines are associated with the development of neuropathic pain, the time-course of their induction after nerve damage has yet to be elucidated. Two peaks of TNF-α were observed (at 6 h and 5 d) after chronic constriction injury in rats and p38 mitogen-activated kinase, an enzyme associated with activation of TNF-α, was activated in the spinal cord, after 5 h and 3 d [5]. In a study of hyperalgesia after administration of a plantar injection of IL-1β into the skin of the hind-paw, the effect of IL-1β was apparent within 1 min [6]. A comprehensive understanding of the serial changes in these cytokines may provide clues to ways of treating the early phase of neuropathic pain induced by nerve injuries.
The development of RNA interference (RNAi) provides a potential alternative approach to treating chronic pain [7]. In a previous study, siRNA targeting the pain-related cation channel P2X3 was infused intrathecally and decreased mechanical hyperalgesia and allodynia, along with a reduction of P2X3, in the dorsal root ganglion and spinal cord [8]. However, the efficacy of the siRNA targeting TNF-α, IL-6, and IL-1β remains to be investigated.
The primary objective of this study was to characterize the time course of expression of the proinflammatory cytokines TNF-α, IL-6, and IL-1β early after nerve injury. The secondary objective was to determine the ability of siRNA targeting those cytokines to reduce the mechanical allodynia and hyperalgesia induced by L5 spinal nerve transection (SNT).
Materials and Methods
Animals
This study was approved (No. 2010-12-098) by the Institutional Animal Care and Use Committee of the author's hospital and complies with Institute of Laboratory Animal Resources guidelines. Efforts were made to limit distress and to use the minimum number of animals necessary to achieve statistical significance as set forth in the guidelines of the International Association for the Study of Pain. Male Sprague Dawley rats (Orient Bio., Sungnam, Korea) weighing 273-375 g at the start of surgery were used. Rats were housed with a controlled light/dark cycle (lights on between 8:00 a.m. and 8:00 p.m.), an ambient temperature of 21-22℃ and unlimited access to a standard laboratory rat diet and water.
Intrathecal catheter implantation and L5 spinal nerve transection
The rats were chronically implanted with intrathecal catheters according to the established method for lumbar catheterization [9]. The catheters (PE 10, INTRAMEDIC, Clay Adams, Becton Dickinson and Company, Rutherford, NJ, USA) were sterilized by immersion in 70% ethanol and fully flushed with sterile water prior to insertion. The rats were anesthetized with sevoflurane in an O2 carrier (induction, 5-8%; maintenance, 3%). Intrathecal catheters were inserted between the L5 and L6 vertebrae using a catheter-through-needle technique and the tip of the catheter was located at approximately L1. After finishing the experiments, lidocaine (15 µl, 20 mg/ml) was given through the catheters to confirm the intrathecal location. Immediate motor paralysis of the hind part of the animal (within 15 s) lasting for 20-30 min indicated the correct intrathecal location. Intrathecal injections of siRNA were carried out 7 d after catheter implantation.
Unilateral peripheral mononeuropathy was produced according to the method described previously [10]. Briefly, rats were anesthetized with sevoflurane in O2 (induction, 5-8%; maintenance, 3%). A small incision in the skin overlying L5-S1 was made, followed by retraction of the paravertebral musculature from the vertebral transverse processes. The L6 transverse process was partially removed, exposing the L4 and L5 spinal nerves. The L5 spinal nerve was identified, lifted slightly, and transected. The wound was irrigated with saline and closed in two layers with 3-0 polyester sutures (fascial plane) and surgical skin staples.
Administration of siRNA
RNAs were administered as described, with modifications [11]. A cocktail of siRNA simultaneously targeting TNF-α (Silencer® Select siRNA; s128522, Ambion, Austin, TX, USA), IL-6 (Silencer® Select siRNA; s217844, Ambion, Austin, TX, USA) and IL-1b (Silencer® Select siRNA; s127941, Ambion, Austin, TX, USA), as well as a control siRNA (Silencer® Negative Control #1 siRNA; Cat #4635, Ambion, Austin, TX, USA), were prepared immediately prior to administration by mixing the RNA (200 µM) with the transfection reagent, i-Fect™ (Neuromics, Minneapolis, MN, USA), in a ratio of 1 : 5 (w : v). At this ratio, the final RNA/lipid complex concentration was 2 µg in 5 µl for each cytokine siRNA and 6 µg in 15 µl for the control siRNA. The cytokine siRNAs were combined and they and the control siRNA (15 µl each) were delivered to the lumbar region of the spinal cord via the intrathecal catheters. Injections were given daily on 5 consecutive days (-1, 0, 1, 2, 3 d after L5 SNT).
Behavioral tests
The rats were acclimatized to the test environment for 15-20 min on three successive days just before the SNT or sham operation. The tests were performed in a quiet room between the hours of 9:00 and 11:00 A.M. The order of testing was fixed, taking into account the degree of distress conferred by each test on the following one. An electronic version of the Von Frey test (dynamic plantar anesthesiometer, model 37400; Ugo Basile, Milan, Italy) for evaluating mechanical allodynia was performed followed by use of an Analgesy-Meter (Ugo Basile, Comerio, Italy) to assess mechanical hyperalgesia.
The allodynia and hyperalgesia thresholds for the mechanical stimuli applied to the hind paw were examined using the electronic Von Frey test. Each rat was placed in a Plexiglas chamber (28 × 40 × 35 cm, wire mesh floor). Fifteen minutes later, servocontrolled mechanical stimuli (using a pointed metallic filament) were applied to the plantar surface at 5-min intervals, exerting a progressively increasing punctate pressure that reached 30 g within 20 s. The pressure evoked a clear voluntary hindpaw withdrawal response (usually at close to 30 g), which was recorded automatically and taken as the mechanical nociceptive threshold. The mechanical threshold was always assessed three times at each time point to yield the mean value. The rats were habituated to the full procedure for 30 min/d on three consecutive days and the experiments were conducted on the fourth day using only those rats that had responded effectively to the mechanical stimulation (cut-off time 40 s).
Mechanical nociceptive thresholds were evaluated using the Analgesy-Meter. Rats were held gently, and incremental pressure (maximum of 250 g) was applied to the dorsal surface of the ipsilateral hind paw. The pressure required to elicit paw withdrawal, the paw-pressure threshold, was determined.
To present the results as percentages of the maximal possible effect (%MPE), the following equation was used: %MPE = [(TV - BV)/(cut-off - BV)] × 100%. In this equation, TV is the test value and BV the baseline value. The cut-off values for mechanical allodynia and hyperalgesia were 30 and 250 g, respectively.
Tissue collection for quantifying cytokine mRNA and protein
Tissue samples were collected and frozen on dry ice, and stored at -70℃. To isolate mRNA and protein, the samples were thawed at room temperature and homogenized in 1 ml of Trizol reagent (Invitrogen, Carlsbad, CA, USA) and PRO-PREP™ (iNtRON Biotechnology, Seoul, Korea), respectively. Isolations was performed according to the manufacturers' protocols.
Reverse-transcription polymerase chain reaction
RNA was reverse-transcribed using an oligo (dT) 15 primer (PrimeScript 1st Strand cDNA Synthesis Kit; Takara Korea Biomedical Inc., Seoul, Korea) in 20 µl reactions. PCR was performed with 1 µl of the first-strand cDNA solution and 0.5 µM each of the sense and antisense primers, for 30 cycles of denaturation (94℃, 30 s), annealing (0℃, 30 s) and extension (72℃, 30 s) for TNF-α, 30 cycles of denaturation (94℃, 30 s), annealing (0℃, 30 s) and extension (72℃, 30 s) for IL-6, 30 cycles of denaturation (94℃, 30 s), annealing (0℃, 30 s) and extension (72℃, 30 s) for IL-1β, or 30 cycles of denaturation (94℃, 30 s), annealing (0℃, 30 s) and extension (72℃, 30 s) for cyclophilin.
The primers used were: TNF-α (5'-3': forward, GTA GCC CAC GTC GTA GCA AAC; reverse, TGT GGG TGA GGA GCA CAT AGT C; base pair, 195), IL-6 (5'-3': forward, AAG TTT CTC TCC GCA AGA GAC TTC CAG; reverse, AGG CAA ATT TCC TGG TTA TAT CCA GTT; base pair, 325), IL-1β (5'-3': forward, CAC CTT CTT TTC CTT CAT CTT; reverse, GTC GTT GCT TGT CTC TCC TTG TA; base pair, 252), cyclophilin (5'-3': forward, GAC AAA GTT CCA AAG ACA GCA GAA A ; reverse, CTG AGC TAC AGA AGG AAT GGT TTG ; base pair, 478). The products were run on 1.5% agarose gels, stained with ethidium bromide and visualized by ultraviolet (UV) illumination. Digital images of the PCR products were produced under UV light, and the intensities of staining, which have been shown to be directly proportional to the amount of DNA, were determined with a densitometer (VersaDoc Multi-Imaging Analyzer Systems; Bio-Rad, Hercules, CA, USA). The amount of each PCR cytokine product was normalized to that of cyclophilin, whose transcript levels do not change in the contused spinal cord.
Protein estimation by ELISA
Total protein concentrations were determined by the bicinchoninic acid assay and used to adjust the results for sample size. IL-1β, IL-6, and TNF-α were determined with ELISA kits (R&D Systems Inc., Minneapolis, MN, USA). Sample optical densities were read at 450 nm (with the correction wave length set at 570 nm), and cytokine concentrations in cell supernatants were calculated from standard curves.
Immunohistochemical staining
Rats were anaesthetized and perfused with 1,000 units of heparin in 0.9% NaCl solution, followed by 4% paraformaldehyde solution in 0.1 M phosphate buffered saline (PBS, pH 7.4). Lumbar spinal cord sections were harvested and processed as described previously [12]. Separated spinal cords were postfixed for 12-15 h in the above fixative and then cryoprotected for 48 h using 20 and 30% sucrose in 4% paraformaldehyde at 4℃. Spinal cords were sectioned on a cryostat (Leica CM 1850) at 25 µm intervals in the transverse plane and stored at -80℃. Before primary antibody incubation the sections were blocked with 3% normal horse serum in PBS (Gibco-Invitrogen, USA) containing 0.1% tween-20 for 1 hr at room temperature. A monoclonal antibody to OX-42 (1 : 500 working dilution; Chemicon, Temecula, CA, USA) was used to reveal the expression of CR3/CD11b on activated microglia, and a polyclonal antibody to glial fibrillary acidic protein (GFAP) (1 : 1,000 working dilution; Sigma-Aldrich, Inc., St. Louis, MO, USA) was used to label astrocytes. The sections were incubated with primary antibodies overnight at 4℃, followed by horse anti-mouse IgG (Vector Labs, Burlingame, CA, USA) for 1hr at room temperature. After multiple rinses in PBS containing 0.1% tween-20, they were incubated with avidin-biotin peroxidase complex (Vector Labs, Burlingame, CA, USA) for 30min at room temperature. The color reaction was stopped at the appropriate time to obtain the brownish color of 3,3'-diaminobensidine (DAB) (Vector Labs, Burlingame, CA, USA) and the sections were rinsed in distilled water to stop the reaction. They were finally counterstained with Hematoxylin QS (Vector Labs, Burlingame, CA, USA), dehydrated in alcohol, cleared in xylene, and covered with coverslips. Control sections were processed as above but without incubation with primary antibodies.
Statistical analysis
Values are expressed as means ± SD or medians (range) as appropriate. The concentrations of cytokines for the two groups at each time point in the in vitro experiment were tested using one-way ANOVA followed by post hoc comparisons (Tukey's post hoc test). The behavioral data for the CON and COCK groups at each time point, were compared using Student's t-test or the Mann-Whitney U test and SigmaStat 3.5 for Windows (Systat Software, Inc., Chicago, IL, USA). The corresponding data on cytokine mRNA levels were analyzed with the Mann-Whitney rank sum test. A value of P < 0.05 was considered significant.
Results
Experimental scheme
To study the time course of expression of the proinflammatory cytokines (TNF-α, IL-6, and IL-1β) after L5 SNT and the effects of the cytokine siRNAs, 80 rats were given control scrambled siRNA (CON group, n = 80) and 70 received the pooled cytokine siRNAs (COCK group, n = 70). Six µg of each siRNA preparation was given via intrathecal catheter 1 d prior to SNT, on the operation day, and 1, 2 and 3 d postoperatively. The levels of the cytokine mRNAs (n = 7, at each time point) and the activation of glial cells, as shown by immunohistochemical staining (n = 3, at each time point) were determined 4, 8 and 12 h, and 1, 2, 4 and 6 d after L5 SNT. Behavioral tests were performed 1 d prior to SNT, and 1, 2, 4 and 6 d after L5 SNT. The scheme of the experiments is summarized in Fig. 1.
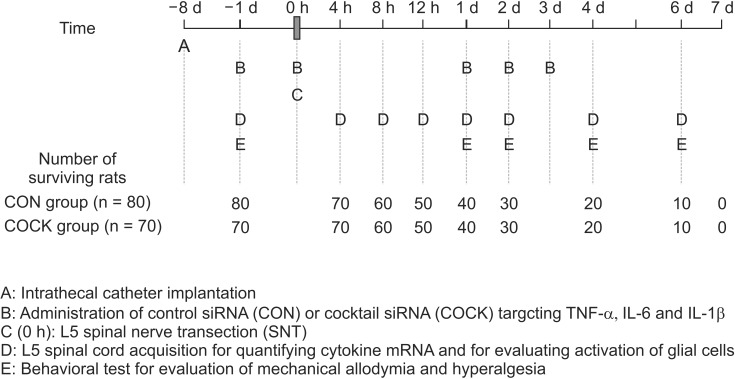
Schematic representation of the experimental procedure used in this study. siRNA: small interfering RNA, CON group: rats allocated to receive control siRNA with a scrambled nucleotide sequence, COCK group: rats allocated to receive the cocktail of siRNAs together targeting TNF-α, IL-6 and IL-1b. In the CON group, cytokine mRNA expressions (n = 7) and the activation of glial cells (n = 3) are determined 7 d after intrathecal catheter implantation without any administration of control siRNA or SNT operation. These data are used as baseline values of cytokine mRNA expression and the activation of glial cells.
The cytokine mRNA data in 63 rats (CON: 38, COCK: 25) and the glial cells activation data in 38 rats (CON: 22, COCK: 16) were used to determine the effectiveness of the siRNAs targeting TNF-α, IL-6, and IL-1β. Forty-nine rats (CON: 16, COCK: 33) were excluded from the analysis for one or other of the following reasons: 1) failure of correct insertion of an intrathecal catheter (n = 16, CON: 9, COCK: 7), 2) failure of a complete SNT model (n = 7, CON: 0, COCK: 7), 3) inappropriate acquisition of spinal cord (n = 5, CON: 2, COCK: 3), 4) technical error during sampling analysis (n = 21, CON: 5, COCK: 16).
The changes in mechanically induced allodynia and hyperalgesia in the rats surviving for 6 d after SNT are shown in Fig. 2. Allodynia and hyperalgesia were lower in the COCK group than in the CON group by 2 d after SNT (P < 0.05) and the difference was maintained for the duration of the experiment.
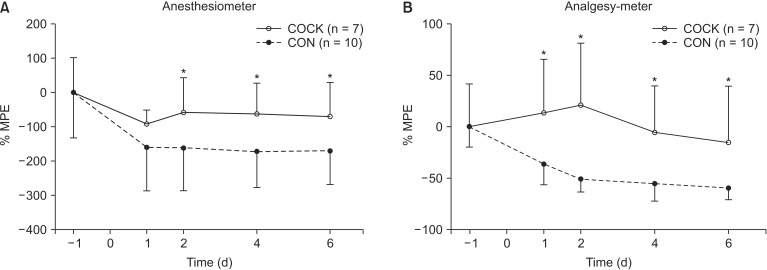
The time course of mechanical allodynia (A) and hyperalgesia (B) in the ipsilateral hind paw of rats undergoing L5 spinal nerve transection (SNT) after the administration of control siRNA (CON group) or a cocktail of small interfering RNAs (siRNA) targeting TNF-α, IL-6 and IL1-β (COCK group). The data on the rats surviving for 6 d after SNT are expressed as mean ± SE. -1: 1 d prior to SNT, 0: the day of L5 SNT, 1, 2, 4 and 6: 1, 2, 4 and 6 d after L5 SNT. *P < 0.05 vs. CON group at each time point. MPE: maximal possible effect. The cut-off values for mechanical allodynia and hyperalgesia are 30 g and 250 g, respectively.
The levels of expression of TNF-α, IL-6, and IL-1β transcripts in the CON and COCK groups are depicted in Figs. 3 and 4, respectively, and the time-course of expression in both groups is shown in Fig. 5. TNF-α level in the CON group increased rapidly after SNT, reaching a maximum (approximately 4.1 fold) at 12 h, and remained high for 6 d (maximum increase ~6.4 fold). IL-6 mRNA in the CON group increased by 12 h after SNT and continued to rise over the 6 d, with considerable variability at 6 d after SNT. A similar increase was observed for IL-1β mRNAs in the CON group. TNF-α and IL-1β mRNA levels at all time points were lower in the COCK group than the CON group (P < 0.05), and the levels of IL-6 mRNA levels at 12 h, 1 and 2 d after L5 SNT in the COCK group were lower than in the CON group (P < 0.05). Overall the administration of siRNA resulted in ~42-64% inhibition of the expression of the proinflammatory cytokines over 6 days.
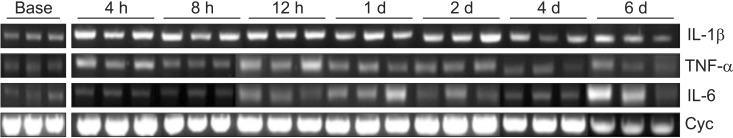
mRNA expression of TNF-α, IL-6, and IL-1β in rats undergoing L5 spinal nerve transection (SNT) (after the administration of control small interfering RNA (siRNA) (CON group). Tissue samples are acquired at 4, 8 and 12 h and 1, 2, 4 and 6 d (n = 3 per each time point) after L5 SNT, and mRNAs for the cytokines are isolated and quantified by reverse transcription polymerase chain reaction (RT-PCR). Cyclophilin is used as a house keeping gene. BASE: 7 d after intrathecal catheter implantation without administration of control siRNA (n = 3). Cyc: Cyclophilin.

mRNA expression of TNF-α, IL-6, and IL-1β in rats undergoing L5 spinal nerve transection (SNT) after the administration of the cocktail of small interfering RNA (siRNA) targeting TNF-α, IL-6 and IL-1β (COCK group). See legend to Fig. 3 for further details. The mRNA levels for TNF-α, IL-6 and IL-1β at 4 h after SNT are not determined because of a technical error which occurred during the analysis process.
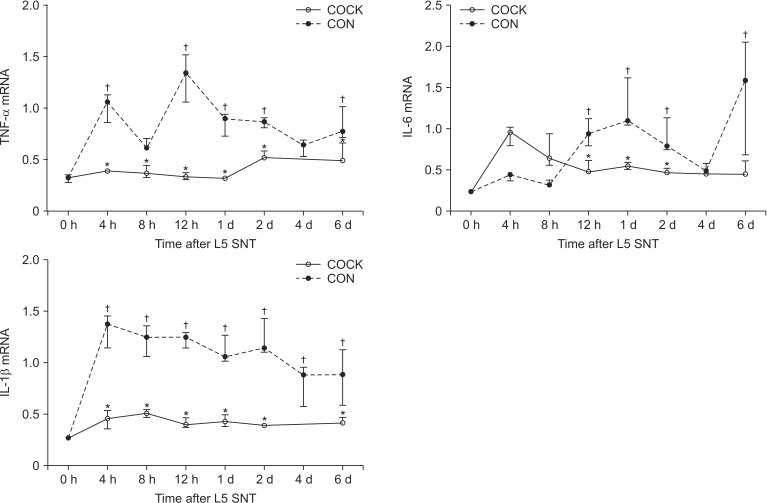
mRNA expression of TNF-α, IL-6 and IL-1β in rats undergoing L5 spinal nerve transection (SNT) and administration of control small interfering RNA (siRNA) (CON group) or the cocktail of siRNA targeting TNF-α, IL-6 and IL-1β (COCK group). Data are expressed as medians (ranges). Cytokine mRNA expression levels are normalized to cyclophilin. 0 h: TNF-α, IL-6 and IL-1β mRNA levels in the spinal cord at 7 d after intrathecal catheter implantation without any administration of control siRNA or the SNT operation are used as the values at time 0 h (n = 3). The cytokine levels at 4 h in the COCK group are not determined because of a technical error during the analysis. *P < 0.05 vs. CON group at each time point. †P < 0.05 vs. 0 h.
The time-course of TNF-α, IL-6 and IL-1β protein levels measured by standard ELISA in the CON group are shown in Fig. 6. The level of IL-6 rose gradually over the 6 d after SNT, while surprisingly those of TNF-α and IL-1β were unchanged.
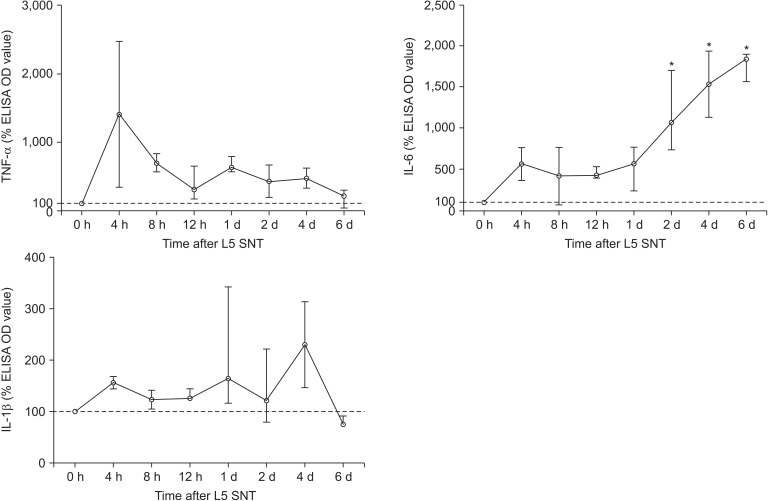
Percentage levels of TNF-α, IL-6 and IL-1β in the rats undergoing L5 spinal nerve transection (SNT) after the administration of the control small interfering RNA (siRNA) (CON group). Data are expressed as the median (range). Each cytokine OD level is normalized to the median OD value at 0 h. The L5 spinal nerve cord samples are harvested on 0, 4, 8, 12 h and 1, 2, 4 and 6 d after L5 SNT to detect these cytokine levels by standard enzyme-linked immunosorbent assay (ELISA). The median OD levels of TNF-α, IL-6 and IL-1β in the spinal cord at 7 d after intrathecal catheter implantation without administration of siRNA or a SNT operation are used as to values. *P < 0.05 vs. 0 h.
The time-course of GFAP and OX-42 expression in the L5 spinal cords of the CON and COCK groups are shown in Figs. 7 and 8, respectively. Taken as a whole, the CON group exhibited greater GFAP immunoreactivity than the COCK group. This was especially clear at 6 d after SNT. Proinflammatory effects were also revealed by enhanced immunostaining for OX-42.
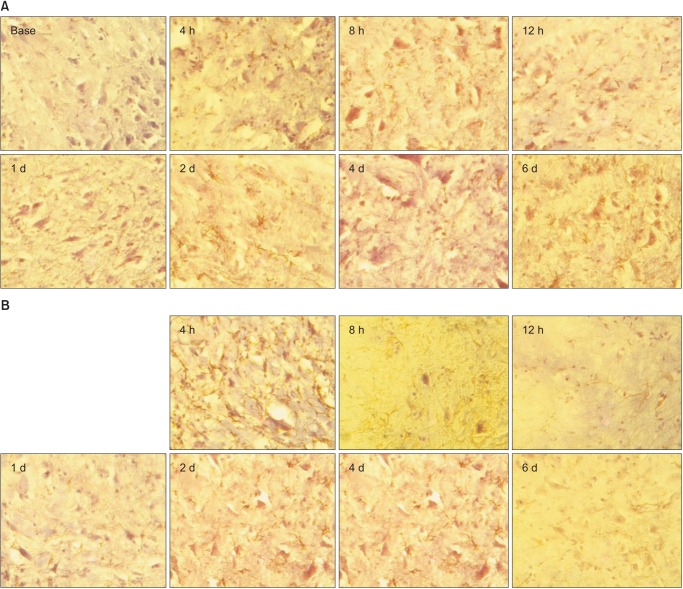
Glial fibrillary acidic protein (GFAP) expression over time in the CON (A) and COCK (B) groups. Astrocytes is determined at 4, 8, 12 h, 1, 2, 4 and 6 d after L5 SNT. BASE: 7 d after intrathecal catheter implantation without administration of siRNA or SNT operation (n = 3 per time point, occasional star-shape astrocytes moderately labeled by GFAP are much more common than in COCK group.
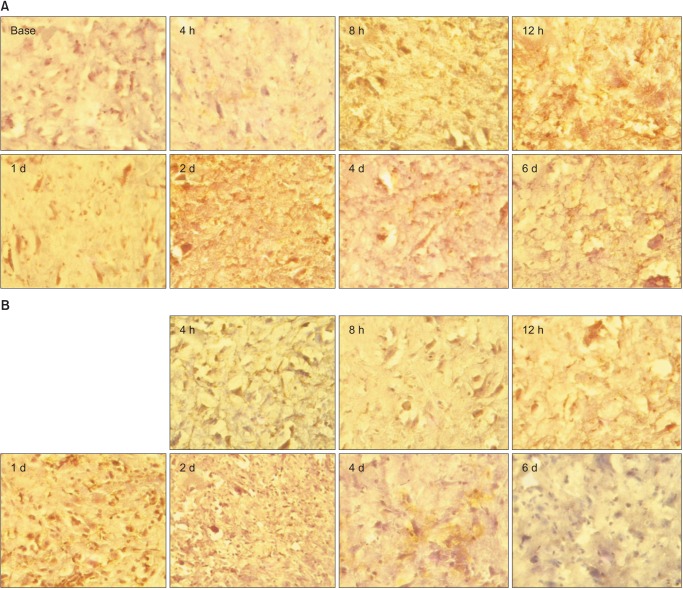
OX-42 expression over time in the CON (A) and COCK (B) groups. Activation of microglial cells by L5 spinal nerve transection (SNT) is determined at 4, 8, 12 h and 1, 2, 4 and 6 d after L5 SNT. BASE: 7 d after intrathecal catheter implantation without any administration of siRNA or SNT operation (n = 3 per time point).
Discussion
The time courses of TNF-α, IL-6 and IL-1β mRNA expression after L5 spinal nerve transection differed somewhat, and siRNAs targeting the three cytokines reduced the mechanical allodynia and hyperalgesia induced by the spinal nerve transection.
Neuropathic pain after nerve injury arises when either the damaged or neighboring undamaged axons are sensitized ectopically such that they induce nociceptive signaling in the central nervous system. Numerous experimental studies have shown that proinflammatory cytokines induce or increase inflammatory as well as neuropathic pain [13,14,15]. Blockade of proinflammatory cytokines such as the three studied here in most cases reduced neuropathic hyperalgesia or allodynia in animal models [13,14,15]. These cytokines are known to contribute to central sensitization [16]. This results in lower thresholds and spontaneous ectopic neuronal firing, which are evident as hyperalgesia and allodynia in the behavioral tests. TNF-α, IL-1β and IL-6 may either directly or indirectly sensitize neurons [16]. For example, IL-1β acts directly on neurons to increase axonal transport of substance P, a potent nociceptive substance, and exogenous TNF-α is able to induce substance P release by sympathetic ganglia [16].
The results of this study may advance our understanding of the effect of TNF-α, IL-1β and IL-6 on neuronal sensitization IL-6 mRNA levels increased somewhat later than those of TNF-α and IL-1β after L5 SNT. TNF-α has a key role in initiating the activation of other cytokines [17]. It exerts its effect through two known receptors, TNF receptor 1 (TNFR1) and TNF receptor 2 (TNFR2). Experimental hyperalgesia has been shown to be dependent on TNFR1 [18] and is mediated via p38 MAPK [14]. In the rat spinal nerve ligation model, intrathecal infusion of the p38 inhibitor SB203580 was only effective if it was started before, not 7 days after SNL, pointing to an important role for TNF-α-dependent signaling early after nerve injury [5]. The peripheral pro-nociceptive action of IL-1β may be mediated by a complex signaling cascade involving the secondary production of nitric oxide, bradykinin and prostaglandins [19]. IL-1β excites nociceptive fibers in vivo within 1 min, suggesting a more direct effect [6]. TNF-α and IL-1 stimulate the production of IL-6 and IL-8 in experimentally-induced inflammatory responses [17]. On the other hand, we observed discordance between mRNA and protein expression in this study (Figs. 5 and 6). The changes in mRNA and protein expression were roughly parallel in the case of IL-6, but not for TNF-α and IL-1β. The failure of protein levels to reflect transcript levels may explained partly by biological and partly by technological considerations. mRNA expression is generally correlated with expression at the protein level for most genes. However, complex biological processes including translational regulation and protein complex formation can affect the expression of specific proteins. No significant correlation between mRNA and protein expression was observed for specific genes in lung adenocarcinomas [20]. Also, our methods of quantifying mRNA and protein were inevitably imperfect.
The crucial role of glial cells in the development of neuropathic pain is known from clinical and experimental studies [2]. Hence, pharmacologic inhibition of glial activation has been proposed as a treatment for neuropathic pain [21]. In the CON group, proinflammation was evident from enhanced immunostaining of both GFAP as a marker of astrocyte and OX-42 as a marker of microglia, which is in accord with previous studies pointing to the importance of glial cells in the development of neuropathic pain.
In 1998, Fire et al. reported that the injection of double stranded RNA (dsRNA) into Caenorhabditis elegans resulted in potent gene silencing [22]. dsRNA several hundred bases in length not only induced significant gene silencing, it was also clearly more active than the corresponding single-stranded antisense molecules, a revolutionary finding in the field of molecular biology. Since the dsRNA molecules interfered with the function of the targeted gene, the process was coined 'RNA interference'. Since then, the popularity of RNAi as a tool for functional genomics has steadily increased. In this process double stranded RNA, is cleaved by the enzyme DICER into 21- to 23-nt duplexes containing 2-nt overhang at each 3' end. These duplexes are incorporated into a protein complex called the RNA-induced silencing complex (RISC). RISC recognizes and cleaves the target mRNA under the direction of the antisense strand of the duplex [7]. Intra-peritoneal injection of siRNA targeting TNF-α inhibited LPS-induced TNF-α gene expression in mice, and the development of sepsis induced by injection of a lethal dose of LPS was significantly inhibited by pre-treatment with anti-TNF-α siRNA [22]. Despite the crucial role of TNF-α, IL-6 and IL-1β in the development of neuropathic pain, little is known about the effects of siRNAs targeting TNF-α, IL-6 or IL-1β. We found that siRNAs simultaneously targeting TNF-α, IL-6 and IL-1β attenuated the mechanical allodynia and hyperalgesia induced by L5 spinal nerve transection. This finding is consistent with the results of Raghavendra et al. [23], who evaluated the effect of inhibition of TNF-α, IL-6 and IL-1β on the acute analgesic effect of morphine, the development of morphine tolerance and opioid withdrawal-induced hyperalgesia in L5 nerve-transected rats. The rats received either saline or morphine and were treated once daily with a cocktail consisting of a fixed-dose combination of an IL-1 receptor antagonist, soluble TNF receptor and goat anti-rat IL-6-neutralizing antibody [23]. Inhibition of these proinflammatory cytokines in the spine reversed the acute morphine antinociception in the nerve-injured rats and also significantly reduced the development of morphine tolerance and withdrawal-induced hyperalgesia and allodynia.
The ease of siRNA delivery is partly dependent on the accessibility of the target organ or tissue within the body. Localized siRNA delivery, which involves the application of siRNA directly to the target tissue, offers several benefits, including better bioavailability and a lessening of the adverse effects typically associated with systemic administration [24]. Intrathecal administration of siRNA was shown to be a good delivery method for evaluating the efficacy of siRNA in rats with neuropathic pain [8,11].
There are certain limitations of this study which should be taken into consideration. The results in the CON group may not reflect accurately the natural time-course of TNF-α, IL-6 and IL-1β after L5 spinal nerve transection in rats. RNAi may result in artificial dysregulation of non-target genes, the results of which are called "off-target effects" [25]. Off-target effects are mainly accounted for by two mechanisms. One is the induction of the interferon response in mammalian cells after transfection with RNA molecules, and the other is the unintended targeting of genes that having sequence homology to the RNA molecule [26]. In general, unwanted effects, such as the activation of the interferon response, are more likely to occur at high siRNA concentrations [25,27]. In in vitro studies, siRNA concentrations of less than 20 nM usually do not lead to any obvious induction of the interferon response [27], although one earlier study reported off-target effects at an siRNA concentration of 10 nM [28]. In this study, such off-target effects may have exerted an influence on the time course of the cytokine mRNA expression, because the concentration of siRNA administered by intrathecal route in the CON group was 200 nM. However, an identical concentration of siRNA was also administered in the COCK group, and the cytokine mRNA levels differed markedly between the CON and COCK groups. Also, we observed no behavioral signs of toxicity induced by off-target effects of the control siRNA. Thus, although the results of this study may not strictly describe the time-dependent characteristics of cytokines after nerve injury, it does appear that the overall characteristics are represented. Additional data, such as the levels of TNF-α, IL-6 and IL-1β in the spinal cord after L5 spinal nerve transection or sham operation without any administration of control siRNA, would help to understand the natural time course of cytokines after nerve injury.
At this time, our results cannot be used clinically to manage neuropathic pain, because most patients who come to pain clinics are considered to be in the chronic stage. However, our results may be useful for developing new treatments for the early phase of neuropathic pain.
In summary, the time courses of TNF-α, IL-6 and IL-1β mRNA expression after L5 spinal nerve transection varied with the cytokine. TNF-α and IL-1β mRNA levels increased early after L5 SNT, IL-6 mRNA levels increased later. These results suggest that certain cytokines play a dominant role in the development of neuropathic pain. RNA interference against these cytokines may prove to be an effective method of attenuating mechanical allodynia and hyperalgesia induced by nerve injuries.
Notes
This is a thesis for a Doctor's degree by Byung Moon Choi.